1.0 - Dynamic All-Pairs Shortest Paths Algorithms
Want to find the shortest path between all pairs of nodes in a graph.
- DIjkstra’s algorithm finds all shortest paths from one node
- The fastest implementation is
- Running Dijkstra’s Algorithm from all nodes is therefore , however this cannot handle negative-weight edges.
- Bellman-Ford is so with an extra loop for each nodes gives
- We can get and handle negative-weight edges for all-pairs
1.1 - Recursive Definition of All-Pairs
-
To solve this problem, we first define some notation
- There are vertices, with IDs
- Uses an adjacency-matrix representation, with as the weight of the edge from vertex to vertex
- If no edge exists, then the weight is , hence return as the weight of the shortest path if one does not exist
-
A path from vertex to can be either:
-
A path with no edges of weight , e.g. for
-
A path with no more than one edge consisting of:
- A path , from vertex to some vertex , and
- The edge
The weight of such path is
-
-
If the shortest path from to has edges, then
- The path from to must have (at most) edges, and
- It must be the shortest path from to
-
Let repeat the weight of the shortest path from to with at most edges
And hence,
-
Since each sub-problem is described by 3 parameters, we require a 3d matrix to store the solutions
1.1.1 - Naive, Slow Solution
-
We store at in an array
-
Calculate , then from , then from etc
slow_apsp(n) // L[m, i, j] is weight of the shortest path from i to j of at most m edges L = new int[n-1][n][n] // Initialise all elements to infinity. L[1] = weights for m = 2 to n - 1 d = L[m-1] d' = L[m] for i = 1 to n for j = 1 to n for k = 1 to n d'[i, j] = min(d'[i, j], d[i, k] + weight(k, j)) -
This algorithm is which is the same as running the Bellman ford algorithm times.
1.1.2 - Improved Solution
This improved time complexity comes from the realisation that we don’t need to compute all $n-1$ matrices - we can compute the shortest paths containing 4 edges, from the set of shortest paths that contain 2 edges, and so on.This means that we only compute a logarithmic amount of these shortest paths vs .
- We calculate then , using
- Then from
- Then from
faster_apsp(n):
// L[m, i, j] is the weight of the shortest path from i to j of at most m edges
L = new int[2(n-1) - 1][n][n]
L[1] = weights
m = 1
while m < n - 1
d = L[m]
d' = L[2m]
for i = 1 to n
for j = 1 to n
for k = 1 to n
d'[i, j] = min(d'[i, j], d[i, k] + d[k, j])
m = 2m
- Thus, this algorithm runs in time
1.2 - Floyd Warshall Algorithm
-
Why did we define our sub-problem in terms of path length?
-
Instead, we can phrase our sub-problem in terms of which intermediate nodes are in the path
-
Let be the shortest path from to , using only intermediate vertices
- Remember that our vertices are conveniently ordered indices
-
We incrementally add a new node to the intermediate set.
-
Extending to requires checking whether the path formed by:
- Going from and is better than already found.
-
Let be the weight of the shortest path from to , using only intermediate vertices
Store $\text{shortestPath}(i, j, k)$ at $d_{ij}^{(k)}$. That is $d_{ij}^{(k)}$ is the weight of t he shortest path from $i$ to $j$ using only intermediate vertices $1\cdots k$
- This algorithm runs in time.
1.2.1 - Floyd Warshall Algorithm Example
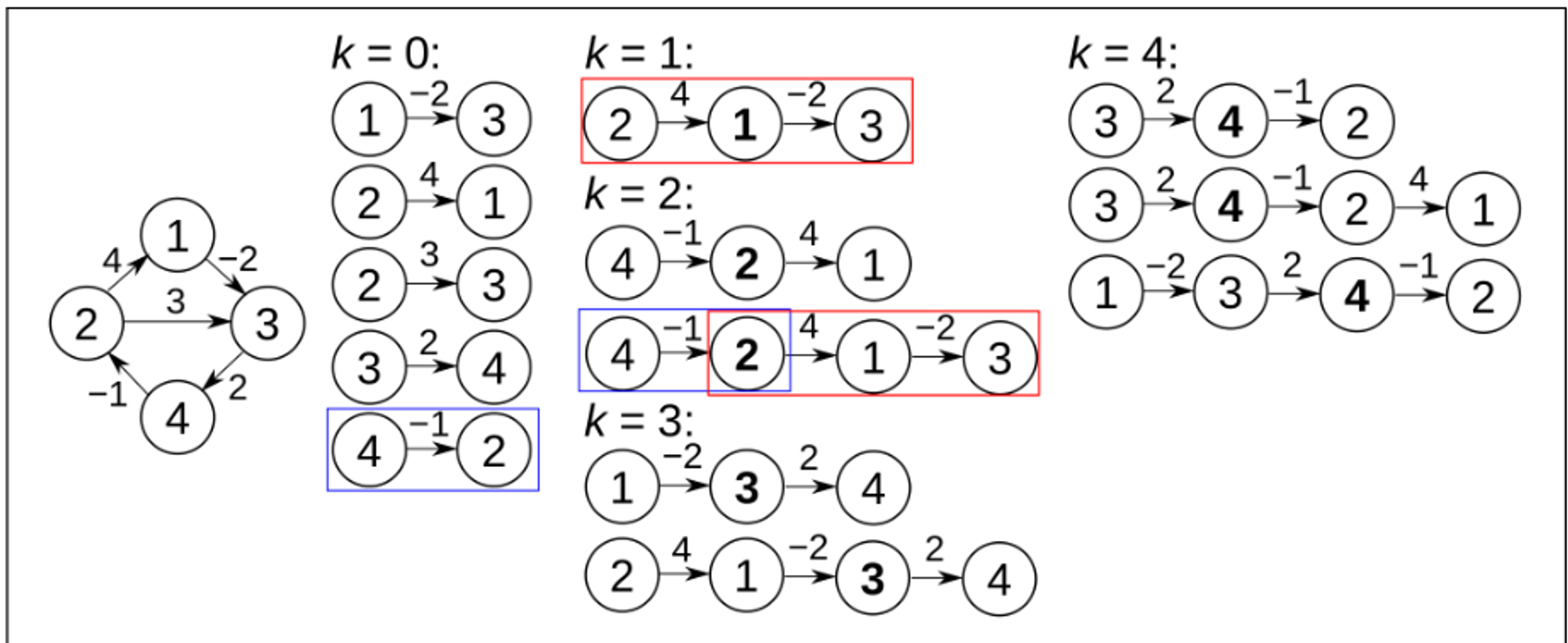
1.3 - Aside: Johnson’s Algorithm
- Johnson’s Algorithm is in the worst case, but for sparse graphs is using an adjacency list representation - in practice this isn’t always faster as it has very large constants associated with it.
- Our strategy is to
- Reweight to eliminate negative-weight edges
- Add a new source vertex.
- Run Dijkstra’s Algorithm for each node